Explainable AI for Neural Networks in Drug Development
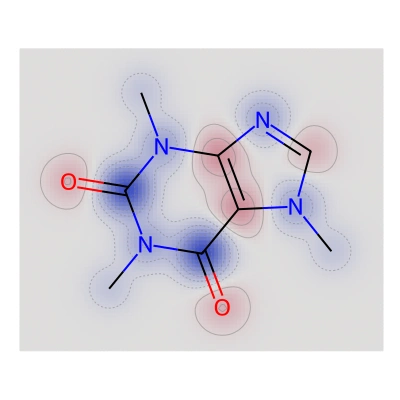 Atom-wise contributions in a caffeine molecule
Atom-wise contributions in a caffeine moleculeIn recent decades, artificial intelligence (AI) and machine learning (ML) have emerged as powerful tools to accelerate drug development, particularly through methods like Quantitative Structure-Activity Relationship (QSAR) modelling, which predict the biological activity of small molecules. However, the inference results of ML models, especially neural networks, are often difficult to interpret for scientists and are frequently referred to as “black boxes”.
To support rational drug design and comply with increasing regulatory demands for model transparency, explainable AI (XAI) methods are now being adopted to provide insights into how ML models make their predictions. In 2024, Koehn AI was approached by a German pharmaceutical company to help integrate the latest advances in XAI techniques into their QSAR neural network Pytorch framework.
Our work focused on evaluating neural-network specific explainability tools such as Integrated Gradients via the Captum library, with more generalizable approaches likes Local Interpretable Model-Agnostic Explanations (LIME) and counterfactual generation using the STONED algorithm in tandem with Self-Referencing Embedded Strings (SELFIES) molecular representations. These methods were benchmarked using real-world activity cliff datasets and evaluated across different model types, including tabular, graph-based and chemical language architectures.
We implemented a robust pipeline that allowed researchers to map predicted molecular activity contributions to individual atoms and molecular fingerprint bits, visualized through RDKit Similarity Maps, offering intuitive, chemically meaningful interpretations. The solution was optimized for performance to ensure seamless and effective integration with the client’s existing workflows and was deployed with their production QSAR platform to support decision making in rational drug design.
We delivered:
- Development of an Explainable AI module integrated into the client’s QSAR neural network framework
- Evaluation of XAI methods including Captum, LIME, and counterfactuals with SELFIES and STONED algorithms
- Support for multiple neural network architectures with visualization with RDKit
- Benchmarking with activity cliff datasets to assess relevance and reliability of explanations
- Optimization of explainability pipelines for efficient end user interaction in workflows
- Deployment of the final module into the client’s production environment supporting informed molecular design